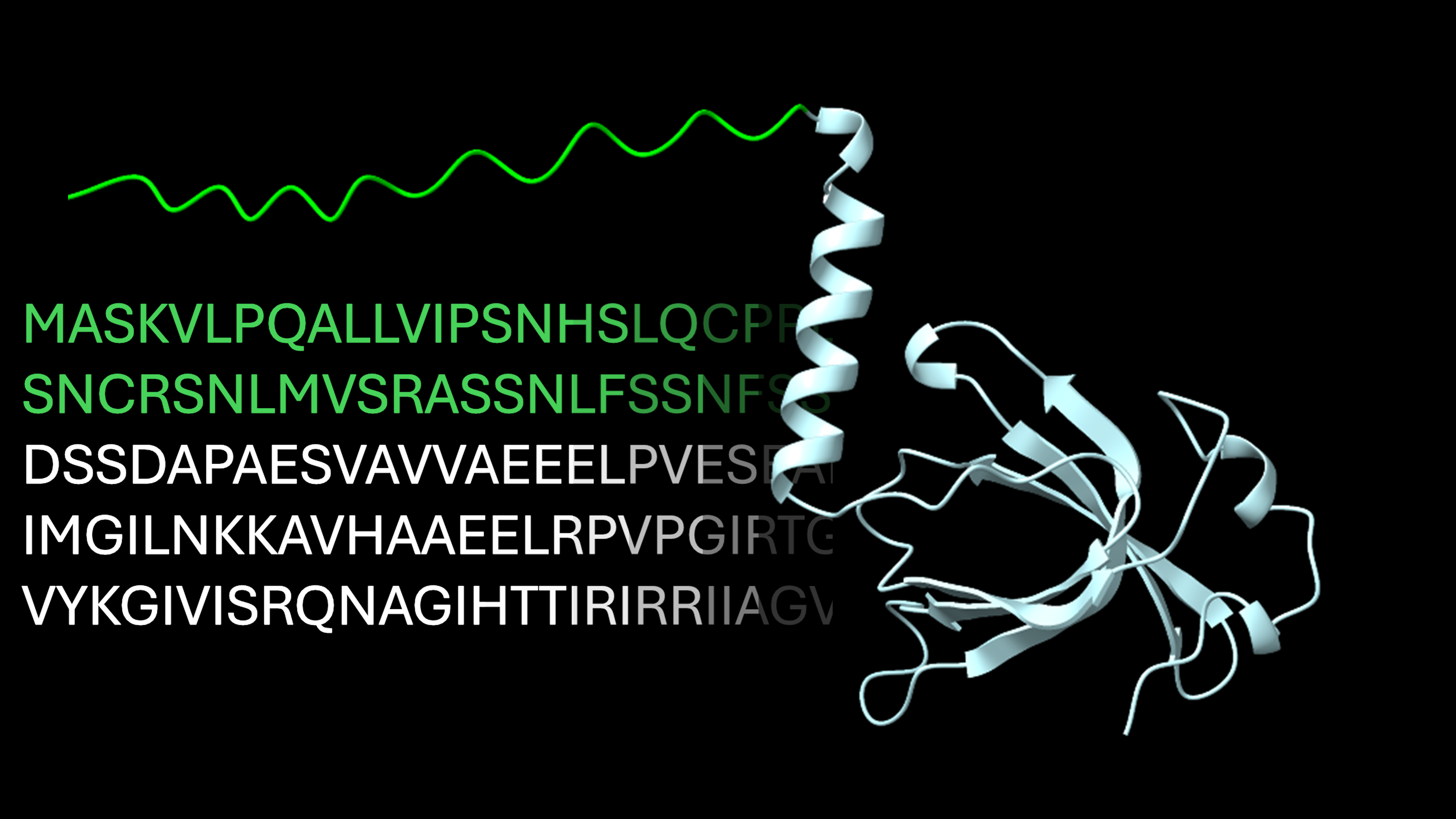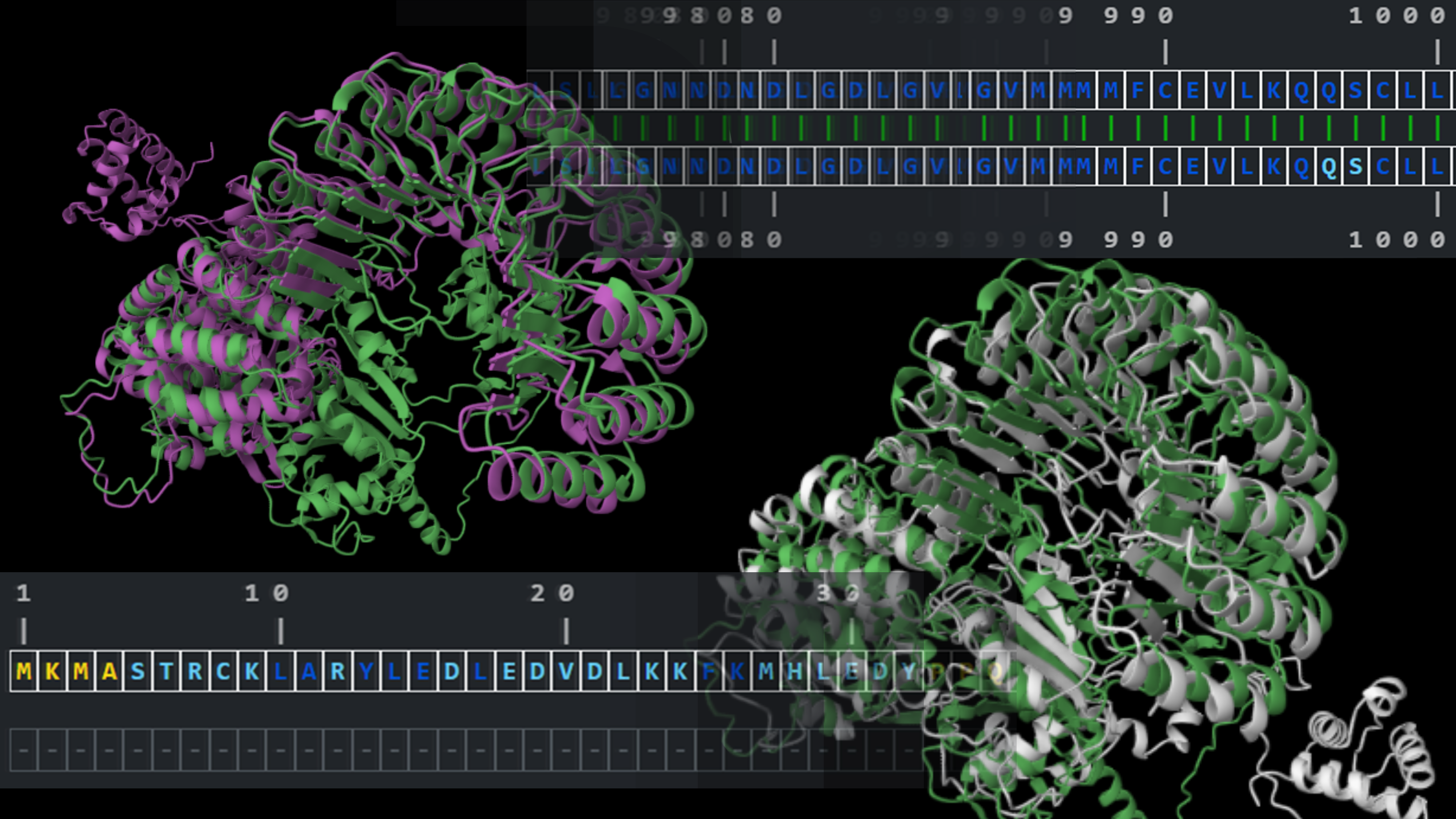
Alpha&ESM hFolds
Comparison of structural models predicted by ESMFold and AlphaFold
E-pRSA
Prediction of residue Relative Solvent Accessibility starting from protein sequence

ISPRED4
Prediction of Interaction Sites starting from protein structure
ISPRED-SEQ
Prediction of Interaction Sites starting from protein sequence
INPS-3D
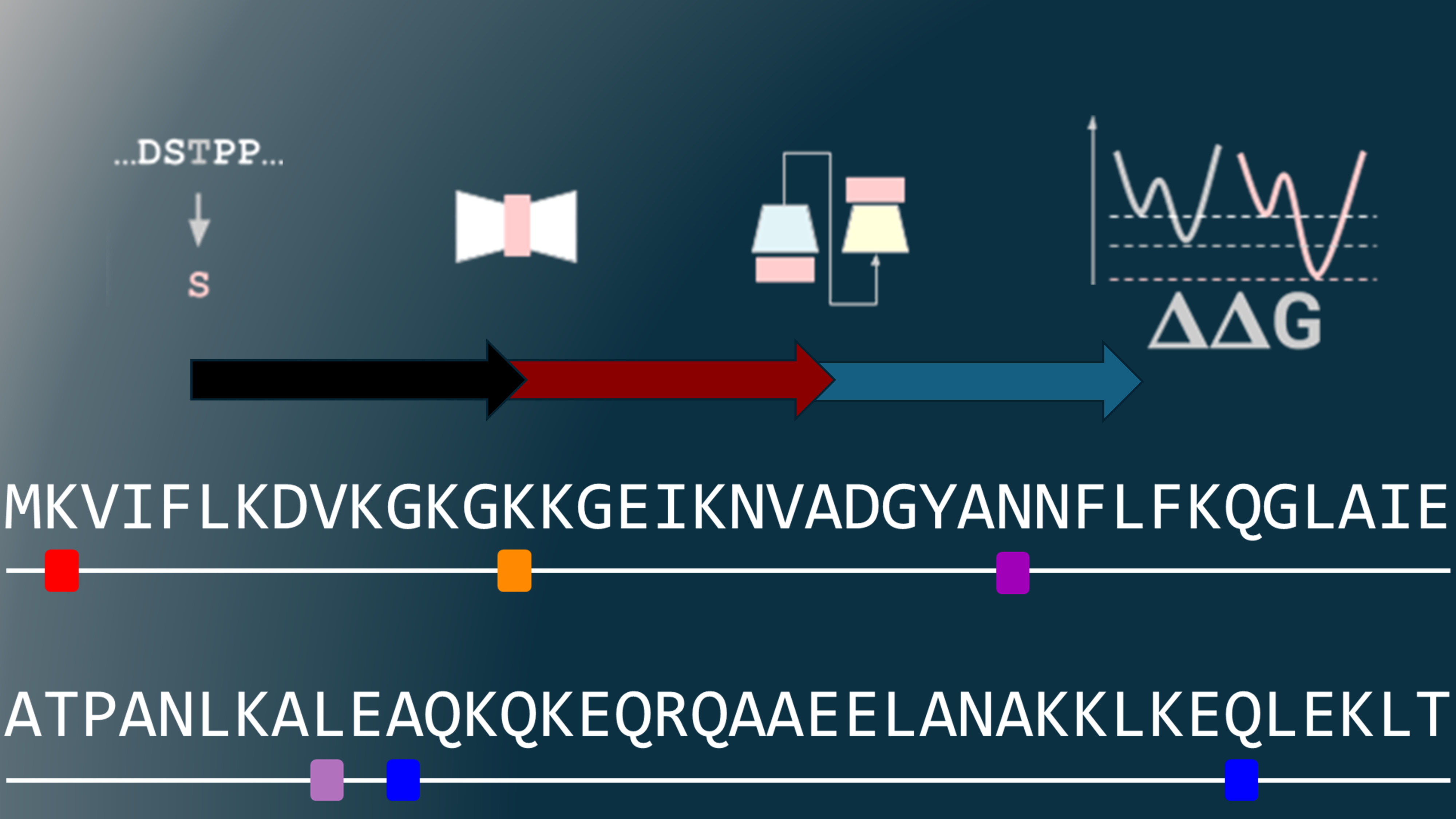
Prediction the impact of protein variants on protein stability starting from protein structure
MultifacetedProtDB
Database providing a comprehensive collection of multifunctional human proteins with annotation