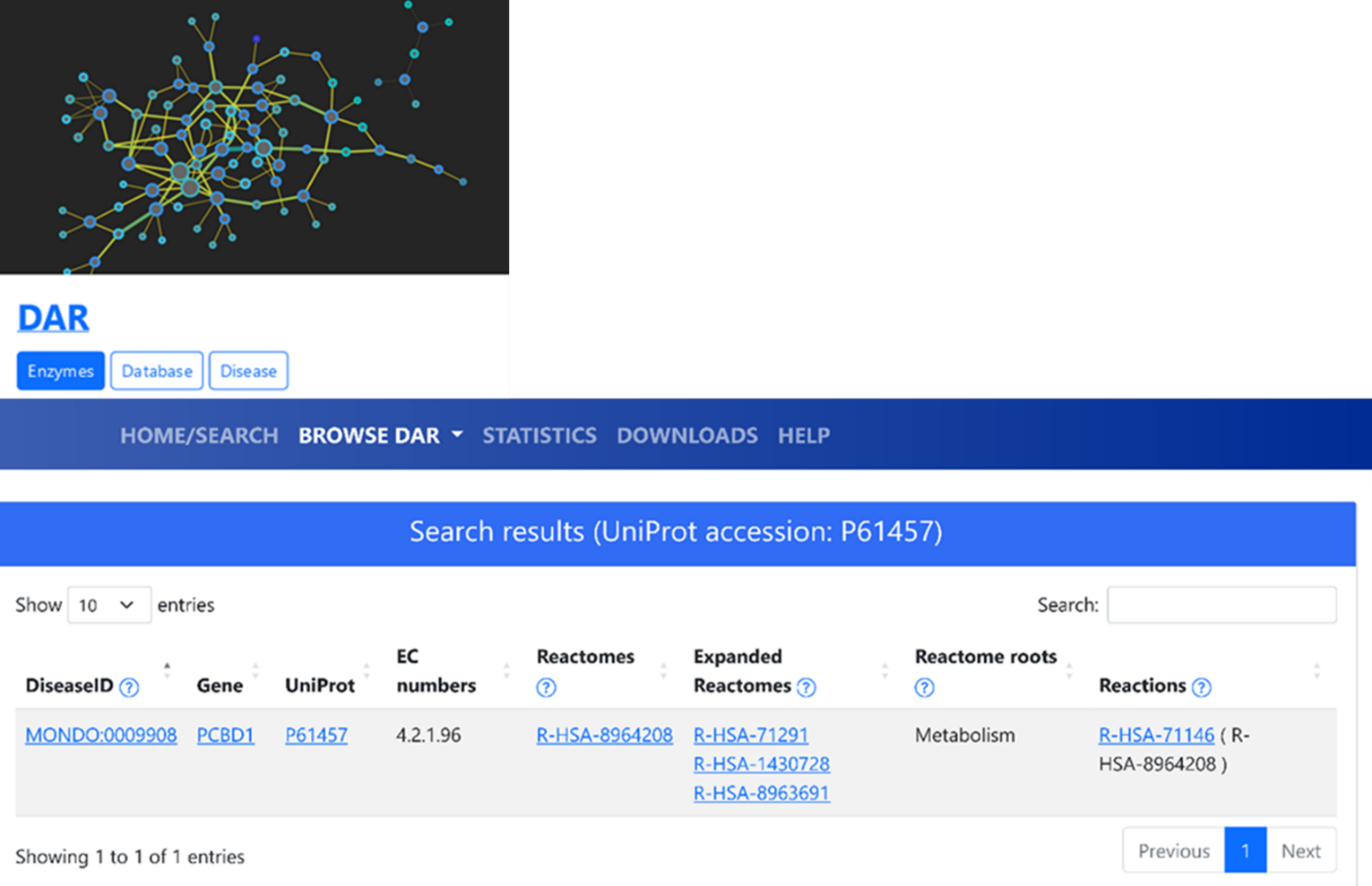
We start our analysis searching the protein in DAR, with the corresponding UniProt accession "P61457". Results reported in Figure 1 show that the protein in an enzyme associated with a single EC number (4.2.1.96), a single disease (MONDO: 0009908, “pterin-4 alpha-carbinolamine dehydratase 1 deficiency”, a benign form of hyperphenylalaninemia due to tetrahydrobiopterin deficiency), and a single Reactome (R-HAS-8964208, “Phenylalanine metabolism”) with one reaction. In the interactive interface of DAR, the names associated to the codes can be retrieved by hovering over the links with the mouse.
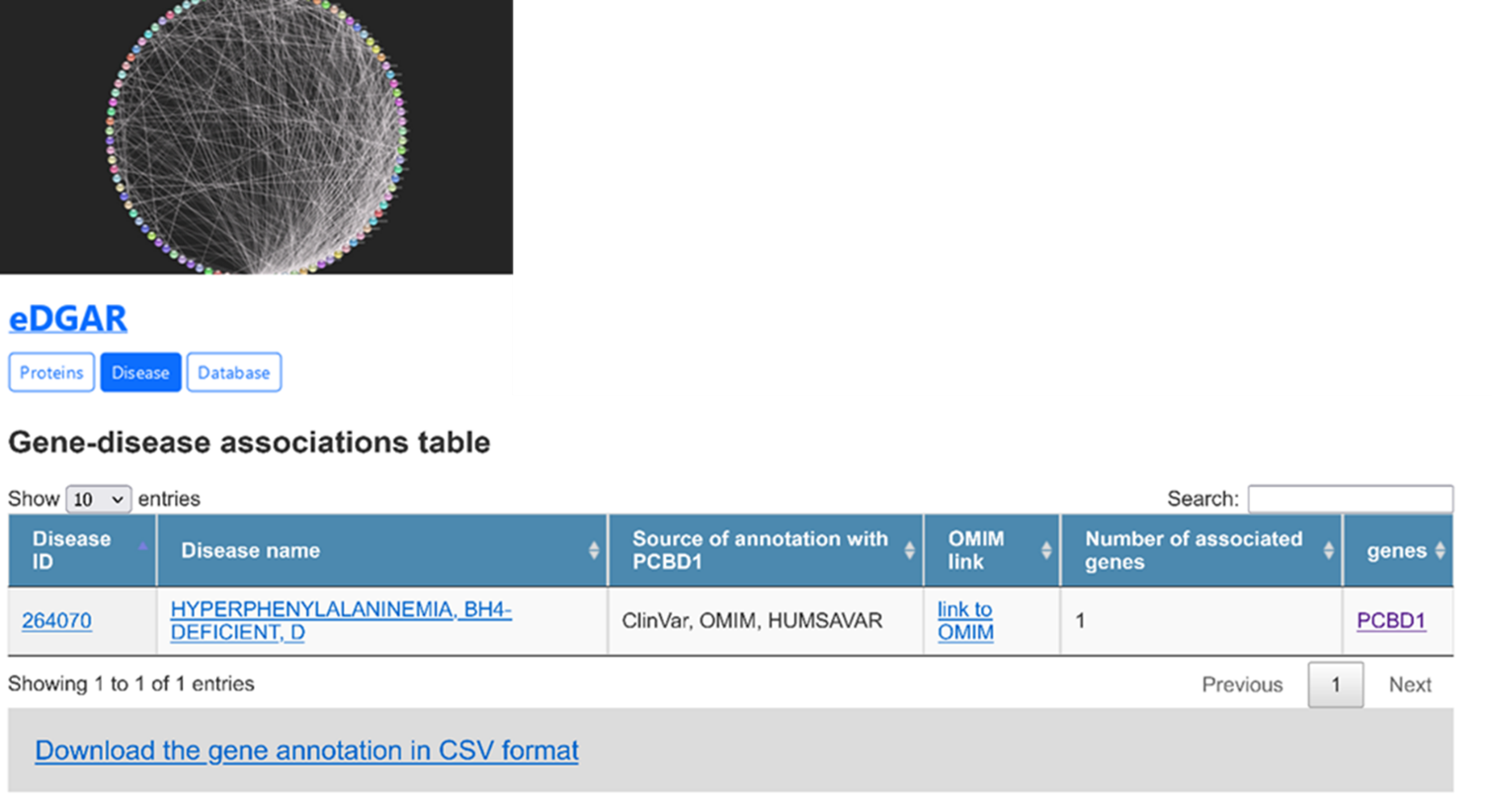
To retrieve more information about the association with diseases, we search eDGAR using the same UniProt accession (Figure 2) and we found again the association with “hyperphenylalaninemia, BH4-deficient, D” (HPABH4D), having OMIM ID "264070".
It is an autosomal recessive disorder characterized by mild transient hyperphenylalaninemia often detected by newborn screening, with increased excretion of 7-biopterin. Patients are almost asymptomatic, although infantile transient neurologic deficits may happen.Patients may also develop hypomagnesemia and nonautoimmune diabetes mellitus during puberty.
Among others, the result page (Figure 2) contains the link to the UniProt webpage, with all protein variants, including the Protein Data Bank (PDB) identifiers of the protein structures.
Links to the Kyoto Encyclopedia of Genes and Genomes (KEGG), to Reactome pathways, Gene Ontology terms, Transcription Factors, and gene annotation, are reported, if available.
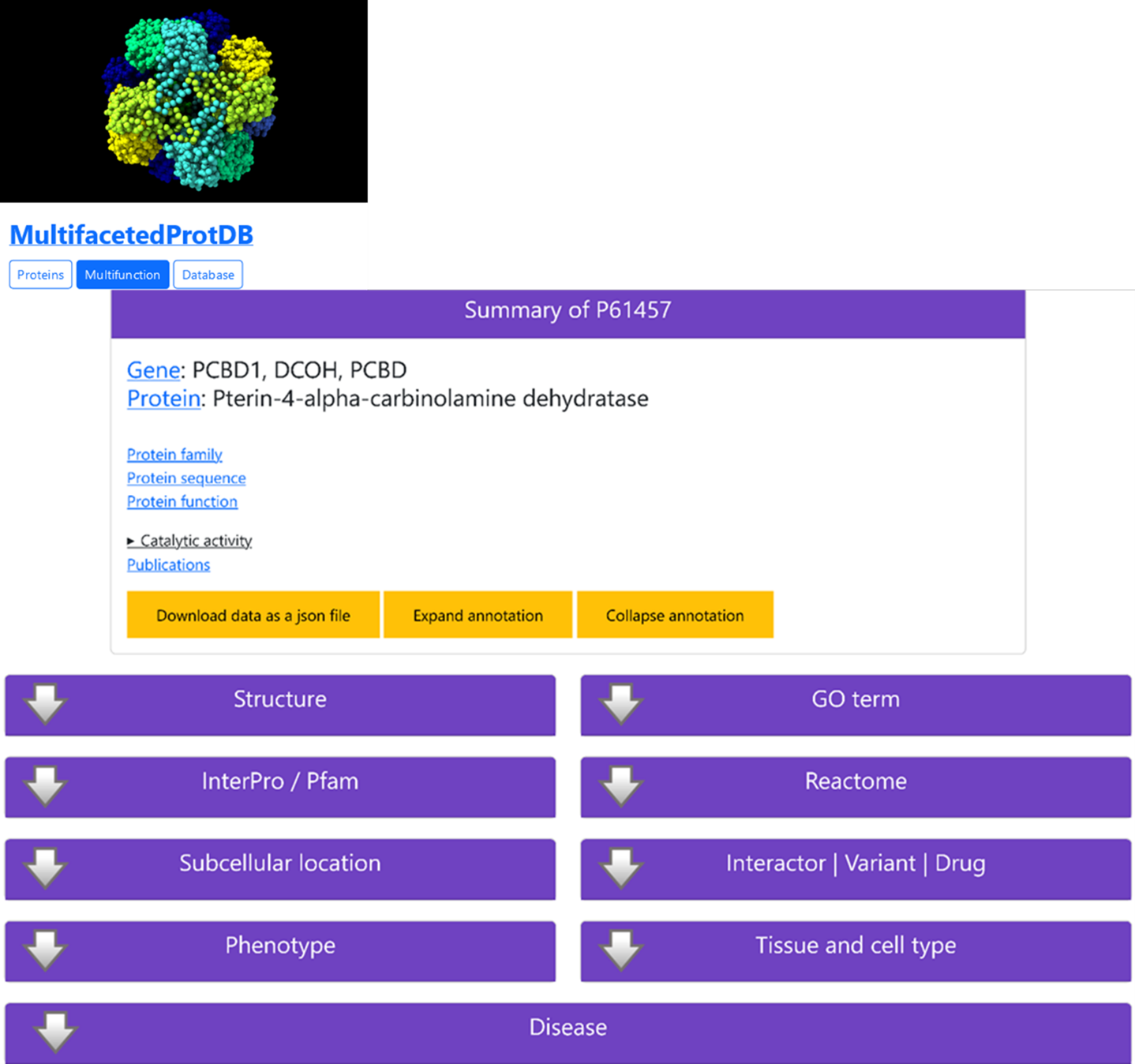
Searching in MultifacetedProtDB indicates that the protein is multifunctional (Figure 3) since it prevents the formation of 7-pterins and accelerate the formation of quinonoid-BH2. It has also been proposed that this protein work as a coactivator for HNF1A-dependent transcription and in the dimerization of homeodomain protein HNF1A, enhancing its transcriptional activity, and it acts as a coactivator for HNF1B-dependent transcription.
The list of all the possible annotation fields found when searching MultifacetedProtDB for gene PCBD1 is in Figure 3.
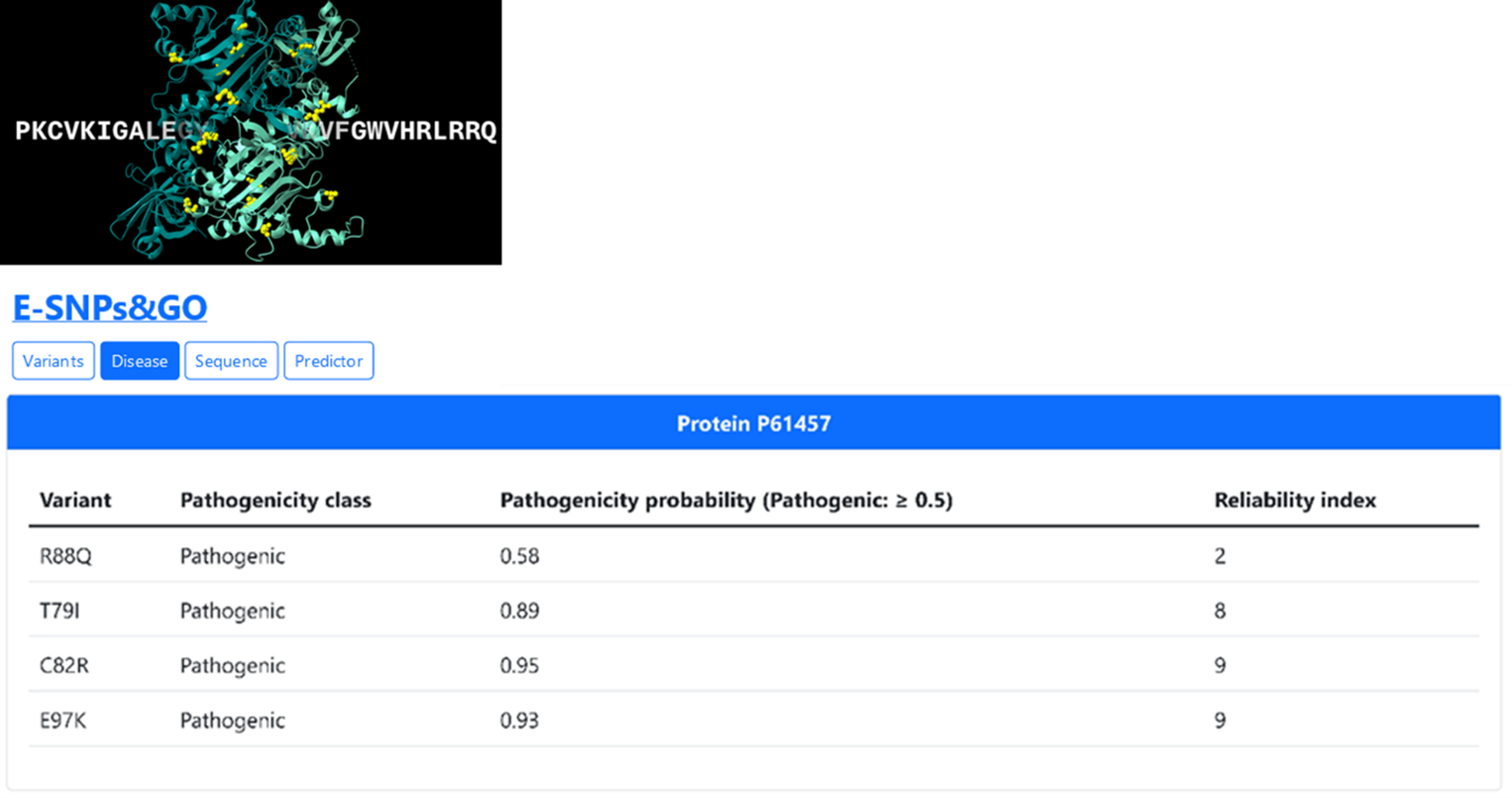
The Variant section of MultifacetedProtDB links the UniProt variant viewer and can be used to collect the pathogenic variants associated to the entry. In this use case, we focus on missense variants associated with the “hyperphenylalaninemia, BH4-deficient, D” (HPABH4D): T79I, C82R, R88Q, E97K.
Finally, we submitted the 4 variants to E-SNPs&GO, that confirms their pathogenicity as reported in the literature (Figure 4).
Summing up we have four variants that are disease related, of a protein whose characteristics are well linked and described in the selected databases.
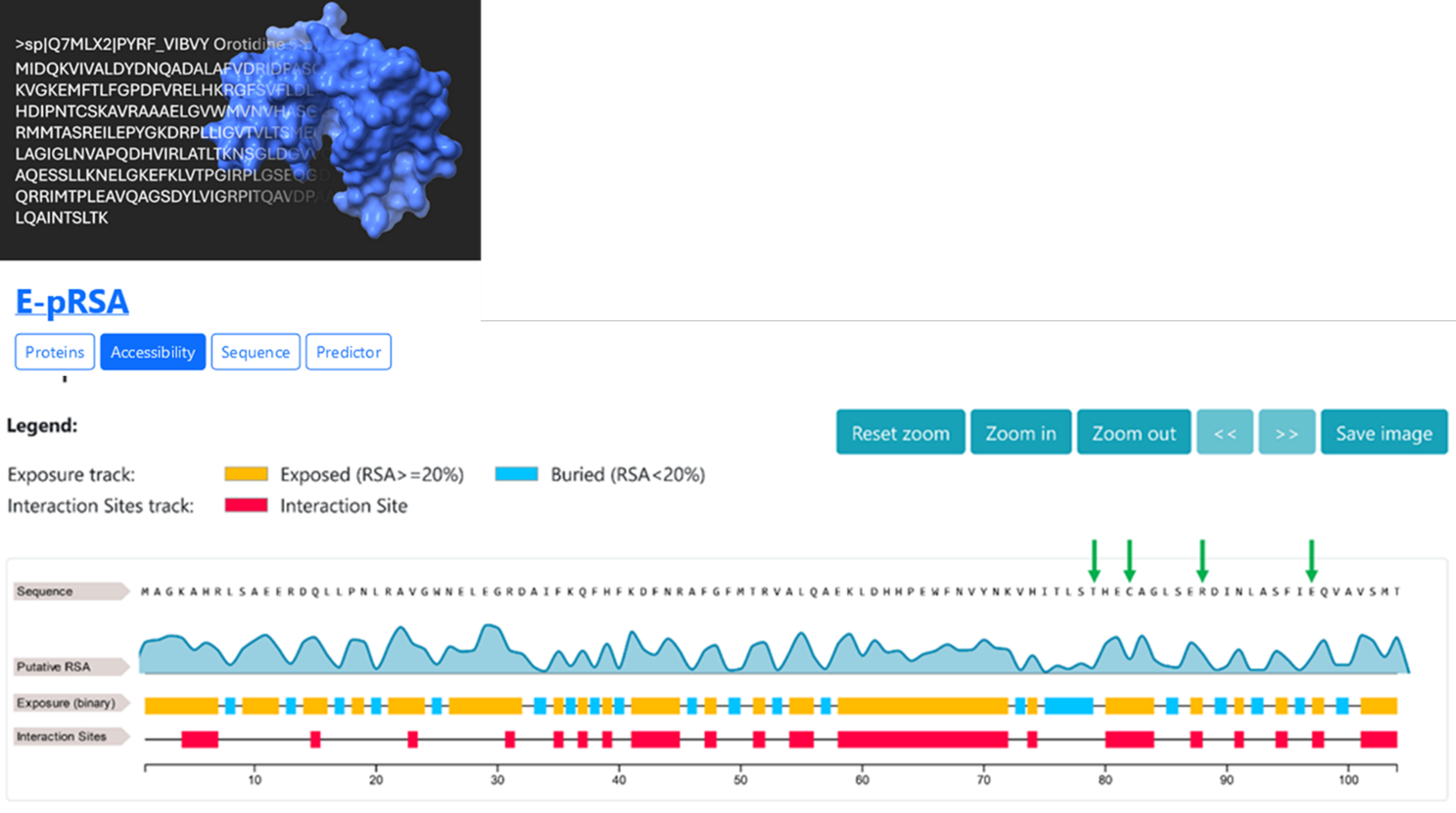
Since the structure of the protein has not been experimentally resolved, we test E-pRSA to predict the Relative Solvent Accessibility (RSA) for the positions of interest with a new encoding procedure called embedding and a convolutional neural network to discriminate the property at hand.
Three out of the four disease related positions, are predicted to be exposed and in interaction sites (Figure 5), while position 79 is predicted to be buried.
We may then investigate the impact of the same variants on protein stability using the INPS (figure 6). It results that all the four variants are predicted to promote a slight destabilization the protein (negative ΔΔG). The effect is more evident for the buried variant T79I (ΔΔG -1.01 kcal/mol).
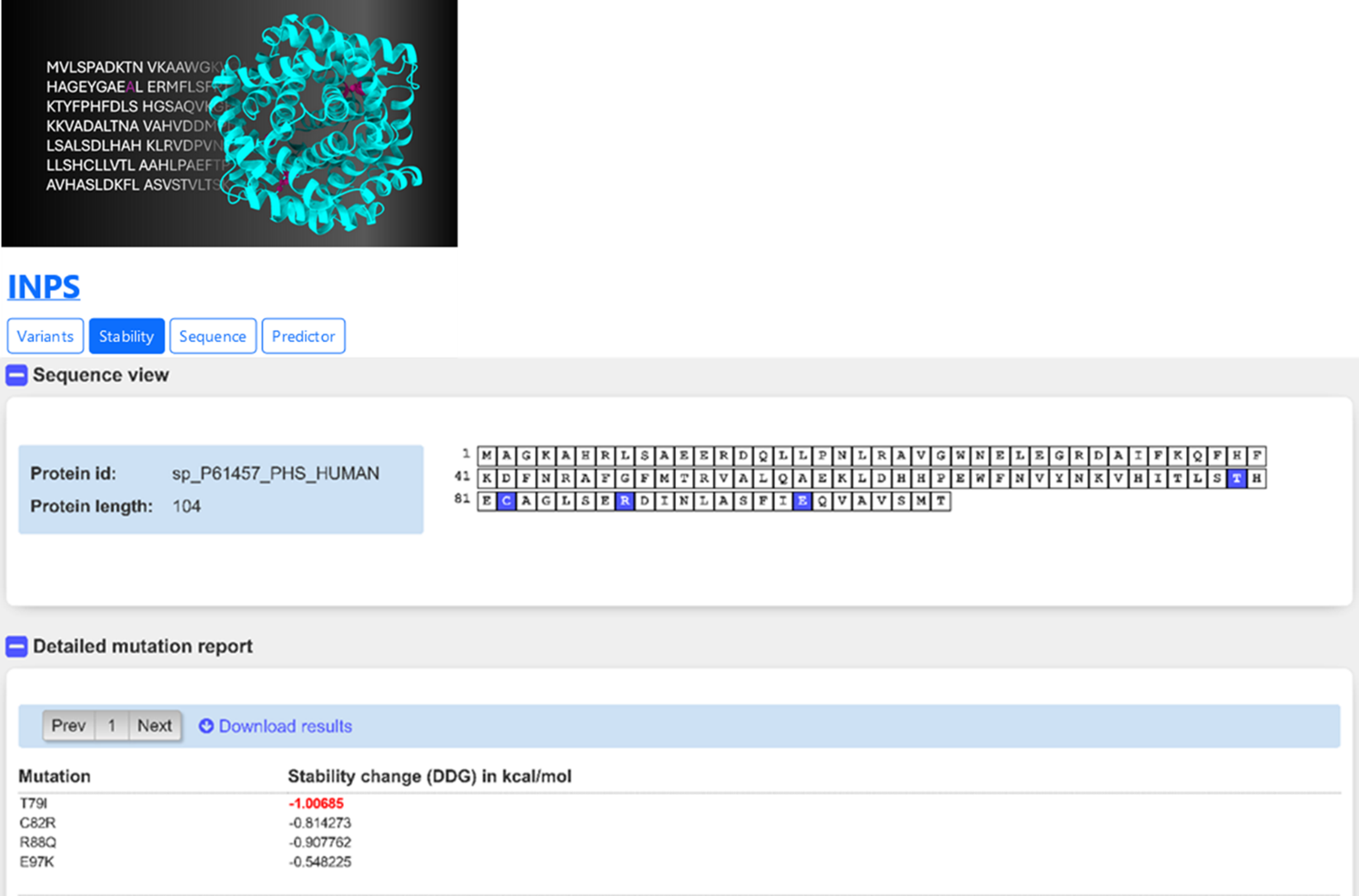
The results, taken together, enable to formulate hypotheses on the molecular effect of the four pathogenic variants of PCDB1. The variant predicted to be buried has a significant impact on the protein stability T79I; the other 3 variants (C82R, R88Q, E97K) are predicted to be only slightly destabilizing. However, they are predicted to be part of a protein-protein interface, which may be altered when they are present. This may alter the protein function and the pathways in which the protein is involved. Indeed, the protein is known to have many interactors. This is so in databases such as BioGRID, IntAct and UniProt, which report for PCDB1 90, 91 and 14 interactors, respectively.